Note: read this paper: stronger LLMs exhibit more cognitive bias. If robust, that would be a very promising result., as I noted in my comment to Kris Wheaton here.

Note: read this paper: stronger LLMs exhibit more cognitive bias. If robust, that would be a very promising result., as I noted in my comment to Kris Wheaton here.

Cyber-security is a broken-window fallacy, but there’s something delightful about this little bot tarpit:

The attacking bot reads the hidden prompt and often traverses the infinite tarpit looking for the good stuff. From Prompt Injection as a Defense Against LLM-drive Cyberattacks (two GMU authors!). HTT Unsupervised Learning (Daniel Miessler)
| ❝ | The modern man thinks that everything ought to be done for the sake of something else, and never for its own sake. |
In praise of idleness, by Bertrand Russel. (HTT Daniel Miessler.)
Adam Russell created the DARPA SCORE replication project. Here he reflects on the importance of Intelligible Failure.
[Advanced Research Projects Agencies] need intelligible failure to learn from the bets they take. And that means evaluating risks taken (or not) and understanding—not merely observing—failures achieved, which requires both brains and guts. That brings me back to the hardest problem in making failure intelligible: ourselves. Perhaps the neologism we really need going forward is for intelligible failure itself—to distinguish it, as a virtue, from the kind of failure that we never want to celebrate: the unintelligible failure, immeasurable, born of sloppiness, carelessness, expediency, low standards, or incompetence, with no way to know how or even if it contributed to real progress.
ADSEI’s Linda McIver on swapping toy problems for real ones:
| ❝ |
I shifted the subject ... teaching the same skills, but now in the context of real datasets, and problems with meaning ... Now they were exclaiming over how useful the skills were. ... solving problems that did not have a textbook solution, so they had to test and verify their solutions. ...
The next year we doubled the number of girls choosing to study the year 11 computer science subject, but we also dramatically increased the number of boys.
|
(My emphasis.)
Mainly I wanted to emphasize that this sensible intervention increased boys' participation as well as girls'.
The take-home seems to be “the reason we haven’t seen [more] progress is that we’ve been teaching it badly”. And that this can be fixed by using real, relevant problems.
Also using STEM is somewhat misleading. Without claiming success, of course women have made notable progress in STEM. But far far less in computer/data science. Which, as McIver notes, has repelled many men as well as women.
If McIver is right, teaching real and relevant problems will stop artificially repelling qualified people.
| ❝ |
When we’re good at participatory sense-making, we can become societies that support each member, where we can each bring as much of ourselves as we like. ...
On our software teams, we have to get good at participatory sense-making to make strong, consistent software. To do this, … We get better at personing. …We get farther together; that frustration has value. ~Jessica Kerr |
Software pushes us to get better as people, on her blog Jessitron.
Jacobs' Heather Wishart interviews Gen. McChrystal on teams and innovation:
I think our mindset should often be that while we don’t know what we’re going to face, we’re going to develop a team that is really good and an industrial base that is fast. We are going to figure it out as it goes, and we’re going to build to need then. Now, that’s terrifying.
On military acquisition:
The mine-resistant vehicles were a classic case; they didn’t exist at all in the US inventory; we produced thousands of them. [after the war started]
This was inspired by Paul Harrison’s (pfh’s) 2021 post, We’ve been doing k-means wrong for more than half a century. Pfh found that the K-means algorithm in his R package put too many clusters in dense areas, resulting in worse fits compared with just cutting a Ward clustering at height (k).
I re-implemented much of pfh’s notebook in Python, and found that Scikit-learn did just fine with k-means++ init, but reproduced the problem using naive init (random restarts). Cross-checking with flexclust, he decided the problem was a bug in the LICORS implementation of k-means++.
Upshot: use either Ward clustering or k-means++ to choose initial clusters. In Python you’re fine with Scikit-learn’s default. But curiously the Kward here ran somewhat faster.
Update Nov-2022: I just searched for the LICORS bug. LICORS hasn’t been maintained since 2013, but it’s popular in part for its implementation of kmeanspp , compared to the default (naive) kmeans in R’s stats package. However, it had a serious bug in the distance matrix computation reported by Bernd Fritzke in Nov. 2021 that likely accounts for the behavior Paul noticed. Apparently fixing that drastically improved its performance.
I’ve just created a pull-request to the LICORS package with that fix. It appears the buggy code was copied verbatim into the motifcluster package. I’ve added a pull request there.
| “ | I believe k-means is an essential tool for summarizing data. It is not simply “clustering”, it is an approximation that provides good coverage of a whole dataset by neither overly concentrating on the most dense regions nor focussing too much on extremes. Maybe this is something our society needs more of. Anyway, we should get it right. ~pfh |
Citation for fastcluster:
Daniel Müllner, fastcluster:Fast Hierarchical, Agglomerative Clustering Routines for R and Python, Journal of Statistical Software 53 (2013), no. 9, 1–18, URL http://www.jstatsoft.org/v53/i09/
Speed depends on many things.
faiss library is 8x faster and 27x more accurate than sklearn, at least on larger datasets like MNIST.I’ll omit the code running the tests. Defined null_fit() , do_fits(), do_splice(),
functions to run fits and then combine results into a dataframe.
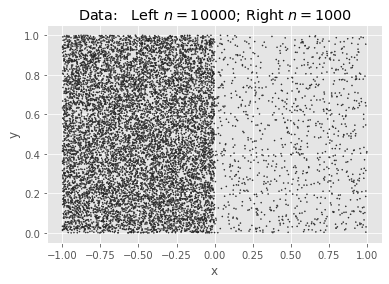
Borrowing an example from pfh, we will generate two squares of uniform density, the first with 10K points and the second with 1K, and find $k=200$ means. Because the points have a ratio of 10:1, we expect the ideal #clusters to be split $\sqrt{10}:1$.
| Name | Score | Wall Time[s] | CPU Time[s] | |
|---|---|---|---|---|
| 0 | KNull | 20.008466 | 0.027320 | 0.257709 |
| 1 | KMeans_full | 14.536896 | 0.616821 | 6.964919 |
| 2 | KMeans_Elkan | 15.171172 | 4.809588 | 69.661552 |
| 3 | KMeans++ | 13.185790 | 4.672390 | 68.037351 |
| 4 | KWard | 13.836546 | 1.694548 | 4.551085 |
| 5 | Polish | 13.108796 | 0.176962 | 2.568561 |
We see the same thing using vanilla k-means (random restarts), but the default k-means++ init overcomes it.
The Data:
SciKit KMeans: Null & Full (Naive init):
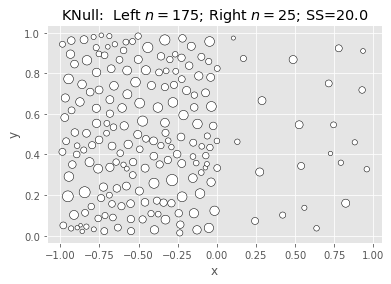
|
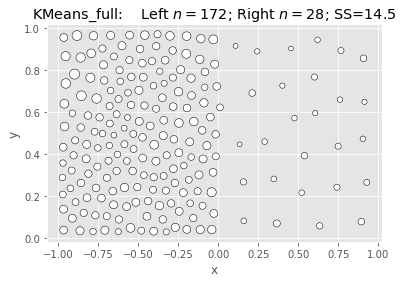
|
SciKit KMeans: Elkan & Kmeans++:
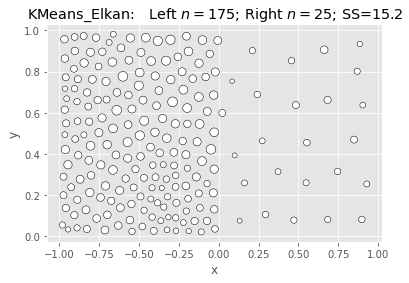
|
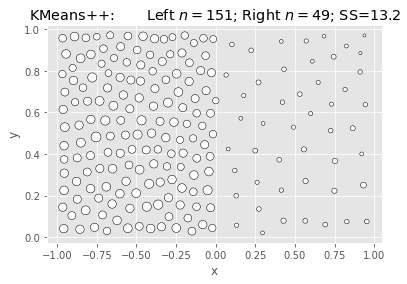
|
Ward & Polish:
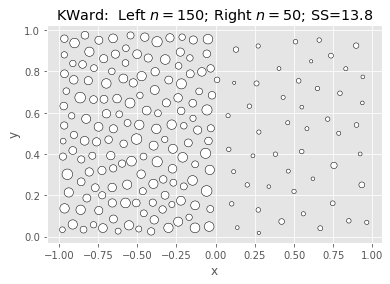
|
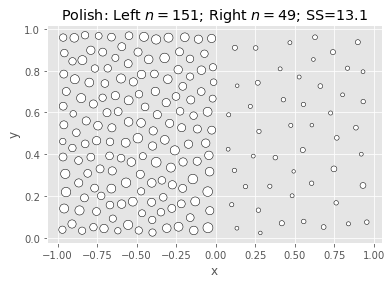
|
What we’re seeing above is that Ward is fast and nearly as good, but not better.
Let’s collect multiple samples, varying $k$ and $n1$ as we go.
| Name | Score | Wall Time[s] | CPU Time[s] | k | n1 | |
|---|---|---|---|---|---|---|
| 0 | KNull | 10.643565 | 0.001678 | 0.002181 | 5 | 10 |
| 1 | KMeans_full | 4.766947 | 0.010092 | 0.010386 | 5 | 10 |
| 2 | KMeans_Elkan | 4.766947 | 0.022314 | 0.022360 | 5 | 10 |
| 3 | KMeans++ | 4.766947 | 0.027672 | 0.027654 | 5 | 10 |
| 4 | KWard | 5.108086 | 0.008825 | 0.009259 | 5 | 10 |
| ... | ... | ... | ... | ... | ... | ... |
| 475 | KMeans_full | 14.737051 | 0.546886 | 6.635604 | 200 | 1000 |
| 476 | KMeans_Elkan | 14.452111 | 6.075230 | 87.329714 | 200 | 1000 |
| 477 | KMeans++ | 13.112620 | 5.592246 | 78.233175 | 200 | 1000 |
| 478 | KWard | 13.729485 | 1.953153 | 4.668957 | 200 | 1000 |
| 479 | Polish | 13.091032 | 0.144555 | 2.160262 | 200 | 1000 |
480 rows × 6 columns
We will see that KWard+Polish is often competitive on score, but seldom better.
Pfh’s example was for $n1 = 1000$ and $k = 200$.
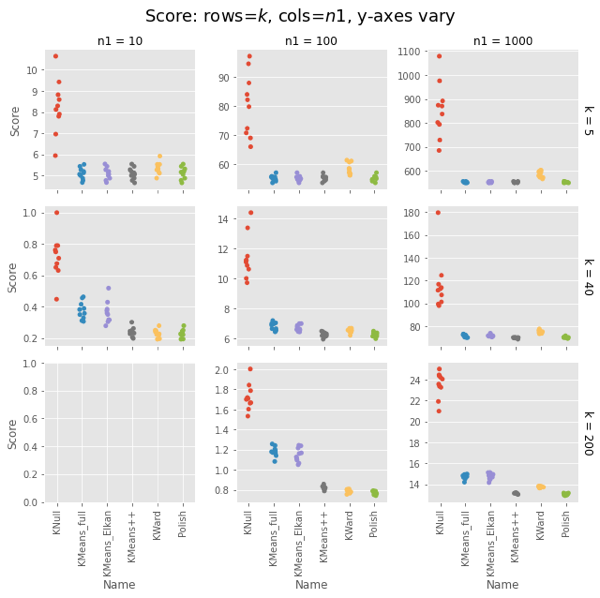
Remember that polish() happens after the Ward clustering, so you should really add those two columns. But in most cases it’s on the order of the KNull.
Even with the polish step, Ward is generally faster, often much faster. The first two has curious exceptions for $n1 = 100, 1000$. I’m tempted to call that setup overhead, except it’s not there for $n1 = 10$, and the charts have different orders of magnitude for the $y$ axis.
Note that the wall-time differences are less extreme, as KMeans() uses concurrent processes. (That helps the polish() step as well, but it usually has few iterations.)
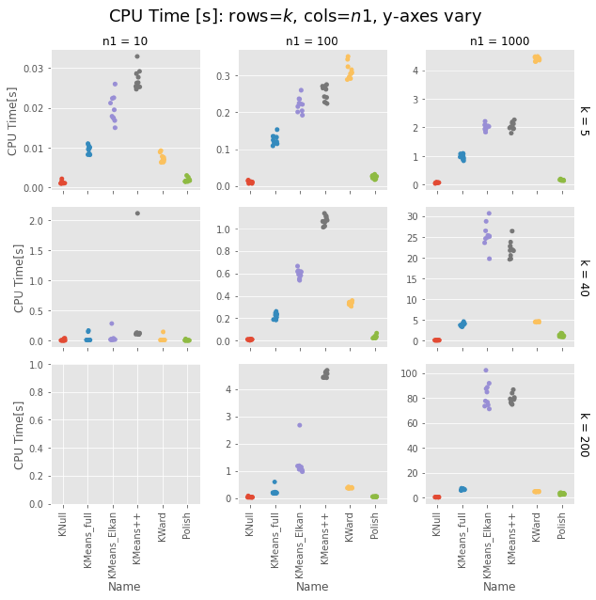
Fair enough: On uniform random data, Ward is as fast as naive K-means and as good as a k-means++ init.
How does it do on data it was designed for? Let’s make some blobs and re-run.
OK, so that’s how it performs if we’re just quanitizing uniform random noise. What about when the data has real clusters? Saw blob generation on the faiss example post.
Preview of some of the data we’re generating:
| Name | Score | Wall Time[s] | CPU Time[s] | k | n1 | |
|---|---|---|---|---|---|---|
| 0 | KNull | 448.832176 | 0.001025 | 0.001025 | 5 | 100 |
| 1 | KMeans_full | 183.422464 | 0.007367 | 0.007343 | 5 | 100 |
| 2 | KMeans_Elkan | 183.422464 | 0.010636 | 0.010637 | 5 | 100 |
| 3 | KMeans++ | 183.422464 | 0.020496 | 0.020731 | 5 | 100 |
| 4 | KWard | 183.422464 | 0.006334 | 0.006710 | 5 | 100 |
| ... | ... | ... | ... | ... | ... | ... |
| 235 | KMeans_full | 3613.805107 | 0.446731 | 5.400928 | 200 | 10000 |
| 236 | KMeans_Elkan | 3604.162116 | 4.658532 | 68.597281 | 200 | 10000 |
| 237 | KMeans++ | 3525.459731 | 4.840202 | 71.138150 | 200 | 10000 |
| 238 | KWard | 3665.501244 | 1.791814 | 4.648277 | 200 | 10000 |
| 239 | Polish | 3499.884487 | 0.144633 | 2.141082 | 200 | 10000 |
240 rows × 6 columns
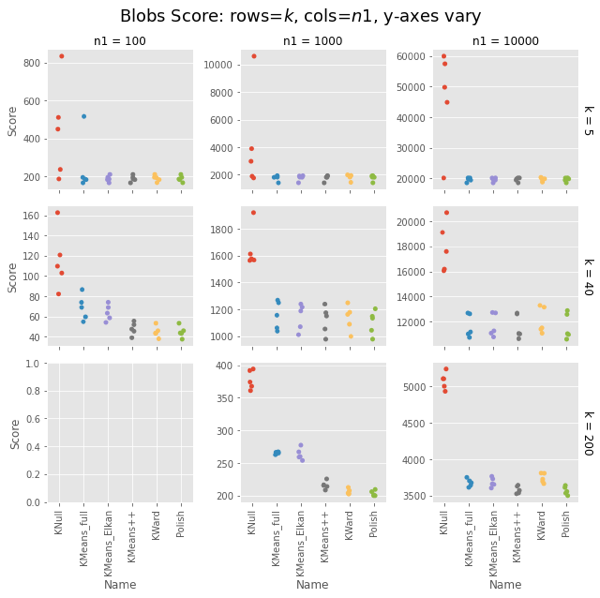
Ward and Polish score on par with KMeans++. For some combinations of $n1$ and $k$ the other algorithms are also on par, but for some they do notably worse.
Ward is constant for a given $n1$, about 4-5s for $n1 = 10,000$. KMeans gets surprisingly slow as $k$ increases, taking 75s vs. Ward’s 4-5s for $k=200$. Even more surprising, Elkan is uniformly slower than full. This could be intepreted vs. compiled, but needs looking into.
The basic result holds.
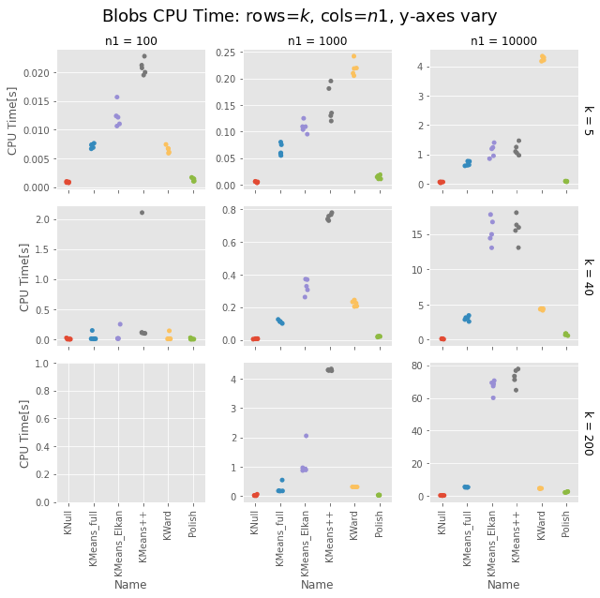
Strangely, Elkan is actually slower than the old EM algorithm, despite having well-organized blobs where the triangle inequality was supposed to help.
On both uniform random data and blob data, KWard+Polish scores like k-means++ while running as fast as vanilla k-means (random restarts).
In uniform data, the polish step seems to be required to match k-means++. In blobs, you can pretty much stop after the initial Ward.
Surprisingly, for sklearn the EM algorithm (algorithm='full') is faster than the default Elkan.
We defined two classes for the tests:
We call the fastcluster package for the actual Ward clustering, and provide a .polish() method to do a few of the usual EM iterations to polish the result.
It gives results comparable to the default k-means++ init, but (oddly) was notably faster for large $k$. This is probably just interpreted versus compiled, but needs some attention. Ward is $O(n^2)$ while k-means++ is $O(kn)$, but Ward was running in 4-5s while scikit-learn’s kmeans ++ was taking notably longer for $k≥10$. For $k=200$ it took 75s. (!!)
The classes are really a means to an end here. The post is probably most interesting as:
KNull just choses $k$ points at random from $X$. We could write that from scratch, but it’s equivalent to calling KMeans with 1 init and 1 iteration. (Besides, I was making a subclassing mistake in KWard, and this minimal subclass let me track it down.)
class KNull(KMeans):
"""KMeans with only 1 iteration: pick $k$ points from $X$."""
def __init__(self,
n_clusters: int=3,
random_state: RandLike=None):
"""Initialize w/1 init, 1 iter."""
super().__init__(n_clusters,
init="random",
n_init=1,
max_iter=1,
random_state=random_state)
Aside: A quick test confirms .inertia_ stores the training .score(mat). Good because it runs about 45x faster.
We make this a subclass of KMeans replacing the fit() method with a call to fastcluster.linkage_vector(X, method='ward') followed by cut_tree(k) to get the initial clusters, and a new polish() method that calls regular KMeans starting from the Ward clusters, and running up to 100 iterations, polishing the fit. (We could put polish() into fit() but this is clearer for testing.
Note: The _vector is a memory-saving variant. The Python fastcluster.linkage*() functions are equivalent to the R fastcluster.hclust*() functions.
class KWard(KMeans):
"""KMeans but use Ward algorithm to find the clusters.
See KMeans for full docs on inherited params.
Parameters
-----------
These work as in KMeans:
n_clusters : int, default=8
verbose : int, default=0
random_state : int, RandomState instance, default=None
THESE ARE IGNORED:
init, n_init, max_iter, tol
copy_x, n_jobs, algorithm
Attributes
-----------
algorithm : "KWard"
Else, populated as per KMeans:
cluster_centers_
labels_
inertia_
n_iter_ ????
Notes
-------
Ward hierarchical clustering repeatedly joins the two most similar points. We then cut the
resulting tree at height $k$. We use the fast O(n^2) implementation in fastcluster.
"""
def __init__(self, n_clusters: int=8, *,
verbose: int=0, random_state=None):
super().__init__(n_clusters,
verbose = verbose,
random_state = random_state)
self.algorithm = "KWard" # TODO: Breaks _check_params()
self.polished_ = False
def fit(self, X: np.array, y=None, sample_weight=None):
"""Find K-means cluster centers using Ward clustering.
Set .labels_, .cluster_centers_, .inertia_.
Set .polished_ = False.
Parameters
----------
X : {array-like, sparse matrix} of shape (n_samples, n_features)
Passed to fc.linkage_vector.
y : Ignored
sample_weight : Ignored
TODO: does fc support weights?
"""
# TODO - add/improve the validation/check steps
X = self._validate_data(X, accept_sparse='csr',
dtype=[np.float64, np.float32],
order='C', copy=self.copy_x,
accept_large_sparse=False)
# Do the clustering. Use pandas for easy add_col, groupby.
hc = fc.linkage_vector(X, method="ward")
dfX = pd.DataFrame(X)
dfX['cluster'] = cut_tree(hc, n_clusters=self.n_clusters)
# Calculate centers, labels, inertia
_ = dfX.groupby('cluster').mean().to_numpy()
self.cluster_centers_ = np.ascontiguousarray(_)
self.labels_ = dfX['cluster']
self.inertia_ = -self.score(X)
# Return the raw Ward clustering assignment
self.polished_ = False
return self
def polish(self, X, max_iter: int=100):
"""Use KMeans to polish the Ward centers. Modifies self!"""
if self.polished_:
print("Already polished. Run .fit() to reset.")
return self
# Do a few iterations
ans = KMeans(self.n_clusters, n_init=1, max_iter=max_iter,
init=self.cluster_centers_)\
.fit(X)
# How far did we move?
𝛥c = np.linalg.norm(self.cluster_centers_ - ans.cluster_centers_)
𝛥s = self.inertia_ - ans.inertia_
print(f" Centers moved by: {𝛥c:8.1f};\n"
f" Score improved by: {𝛥s:8.1f} (>0 good).")
self.labels_ = ans.labels_
self.inertia_ = ans.inertia_
self.cluster_centers_ = ans.cluster_centers_
self.polished_ = True
return self
Usage: KWard(k).fit(X) or KWard(k).fit(X).polish(X).

Slogan on an ad for scrum training. Featured in the the 2015 talk “Agile is Dead”.
Dave Thomas shows what goes wrong when “Agile” becomes a proper noun, and talks about reclaiming agility.
Find out where you are.
Take a small step towards your goal.
Adjust your understanding based on what you learned.
Repeat.
[All things equal], take the path that makes future change easier.
A good column by our SVP for technology & innovation.

Bookmarking A methodology for agile data science by Edwin Thoen (ch.5 of Agile Data Science with R). Also favors Kanban. Highlights below.
As a rule of thumb, never work on more tasks simultaneously than the number of data scientists on the team.
One of the major pitfalls of trying to improve a data science product is endless exploration of a hypothesis.
How to manage exploration then?
The data scientist should not take longer for the task than the team agreed upfront, wrapping up even when he does not feel completely finished. If he found an alleyway that is still worthwhile exploring a new task should be put in the backlog, instead of persevering in the current task
Bookmarking Why Kanban for data science. Two core issues:
…tasks have uncertain outcomes
…don’t have a good definition of done
Wait… @agilelisa, this is near your domain, no?
In Slack and Zoom were distracting…, John Lacy notes:
The set new rules: