Excellent 12-minute presentation & performance by Joy Buolamwini on AI bias. From 2019: youtu.be/_sgji-Bla…
Excellent 12-minute presentation & performance by Joy Buolamwini on AI bias. From 2019: youtu.be/_sgji-Bla…
The mouseover for reviews on “Trusted [hah] Consumer Reviews”:
<img style=“display:block; margin-left:auto; margin-right:auto;” src=“https://cdn.uploads.micro.blog/15308/2021/bfccc9036a.png" alt=“Text screenshot: “Trusted Consumer Reviews … the scoring … should not be used for accuracy purposes. …These referral fees may affect the rankings.”” title=“TrustedHahConsumerReviews.png” border=“0” width=“240” height=“134” />
sigh
Voice for compassionate harm reduction “when truth itself has supply-chain problems.”
| ❝ | It's easy to judge the unvaccinated. As a doctor, I see a better alternative |
In the last couple of years I’ve discovered both Conversations with Tyler and Sullivan’s (new) blog. So I’m looking forward to Tyler’s convo with Sullivan.
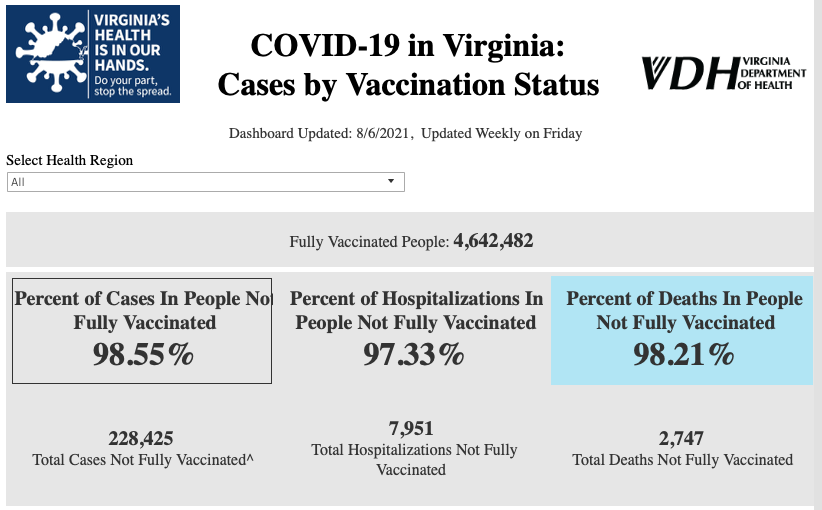
This is 5-10x more dramatic than I thought.
This doesn’t seem to change much if you zoom in to recent months.
🔖 Old-fashioned confusion, a forecasting blog by Foretell forecaster kojif.
🔖 Alan Jacobs' post, beats me. What he said.
On the narrow point, there are risk-benefit analyses – like Peter Godfrey Smith on lockdowns – but like Jacobs and contra Dougherty, that’s not most of what I see. Dougherty seems better on sources of fear.
| ❝ | the reality and importance of climate change does not ... excuse ... avoiding questions of research integrity any more than does the reality and importance of breast cancer. |
I love this Foretell comment:
The Economist often has clever covers, but I really struggled to figure out the metaphor here. Sunblock and shades for China policy? And weird to have brand name product placement.
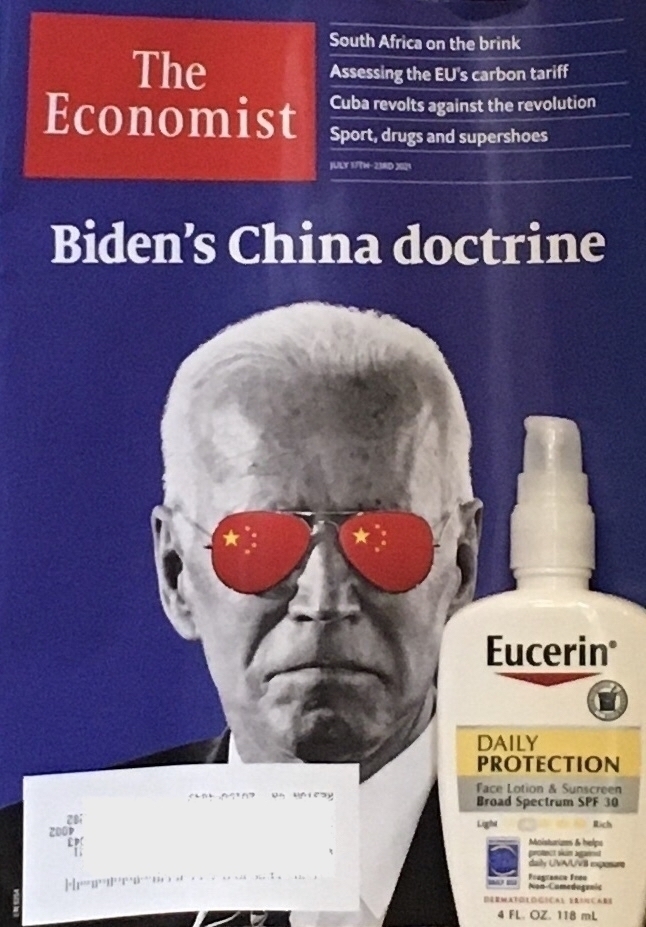
Then on doubletake I realized someone had put a real bottle of sunblock on the stairs next to the mail.
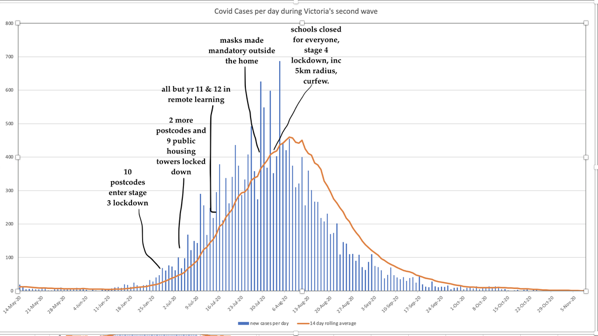
Watching Sydney’s Delta cases repeat the early-phase exponential growth of Melbourne, ADSEI’s Linda McIver asks:
Would our collective understanding of covid have been different if we were all more data literate?
Almost certainly, and I’m all for it. But would that avoid
watching Sydney try all of the “can we avoid really seriously locking down” strategies that we know failed us, … like a cinema audience shouting at the screen,
Not necessarily. Probably not, even, but that’s OK. It would still be a huge step forward to acknowledge the data and decide based on costs, values, and uncertainties. I’m fine with Sydney hypothetically saying,
| ❝ | You're right, it's likely exponential, but we can't justify full lockdown until we hit Melbourne's peak. |
I might be more (or less) cautious. I might care more (or less) about the various tradeoffs. I might make a better (or worse) decision were I in charge. That’s Okay. Even with perfect information, values differ.
It’s even fine to be skeptical of data that doesn’t fit my preferred theory. Sometimes Einstein’s right and the data is wrong.
What’s not okay is denying or ignoring the data just because I don’t like the cost of the implied action. Or, funding decades-long FUD campaigns for the same reason.
PS: Here is Linda’s shout suggesting that (only) stage-4 lockdown suppressed Delta:
Acapella Science’s 🧬 Evo Devo is just so good.
(So many others too.)
The July 5 Lancet letter reaffirms the authors' earlier skeptical view of LabLeak. They cited some new studies and some older pieces in favor of Zoonosis. Summary of those sources below. (This post was originally a CSET-Foretell comment.)
(Limitation: I’m summarizing expert arguments - I can evaluate arguments and statistics, but I have to rely on experts for the core biology. )
Cell July 9: Four novel SARS-CoV-2-related viruses sequenced from samples; RpYN06 is now second after RaTG13, and closest in most of the genome, but farther in spike protein. Also, eco models suggest broad range for bats in Asia, despite most samples coming from a small area of Yunnan. The upshot is that moderate looking found more related strains, implying there are plenty more out there. Also, a wider range for bats suggests there may be populations closer to Wuhan, or to its farm suppliers.
May 12 Virological post by RF Garry: Thinks WHO report has new data favoring zoonotic, namely that the 47 Huanan market cases were all Lineage B, and closely-related consistent with a super-spreader; however, at least some of the 38 other-market cases were Lineage A. Both lineages spread from Wuhan out. Zoonosis posits Lineage A diverged into Lineage B at a wildlife farm or during transport, and both spread to different markets/humans. LabLeak posits Lineage A in the lab, diverging either there or during/after escape. Garry thinks it’s harder to account for different strains specifically in different markets. And the linking of early cases to the markets, just like the earlier SARS-CoV. A responder argues for direct bat-to-human transfer. Another argues that cryptic human spread, only noticed after market super-spread events, renders the data compatible with either theory.
Older (Feb) Nature: Sequenced 5 Thai bats; Bats in colony with RmYN02 have neutralizing antibodies for SARS-CoV-2. Extends geog. distribution of related CoVs to 4800km.
June Nature Explainer: tries to sort un/known. “Most scientists” favor zoonosis, but LL “has not been ruled out”. “Most emerging infectious diseases begin with a spillover from nature” and “not yet any substantial evidence for a lab leak”. Bats are known carriers and RATG13 tags them, but 96% isn’t close enough - a closer relative remains unknown. “Although lab leaks have never caused an epidemic, they have resulted in small outbreaks”. [I sense specious reasoning there from small sample, but to their credit they point out there have been similar escapes that got contained.] They then consider five args for LL: (1) why no host found yet? (2) Coincidence first found next to WIV? (3) Unusual genetic features signal engineering (4) Spreads too well among humans. (5) Samples from the “death mine” bats at WIV may be the source. In each case they argue this might not favor LL, or not much. Mostly decent replies, moving the likelihood ratio of these closer to 1.0, which means 0 evidence either way.
Justin Ling’s piece in FP: I’m not a fan. As I’ve argued elsewhere, it’s uneven at best. Citing it almost count agains them. Still, mixed in with a good dose of straw-man emotional arguments, Ling rallies in the last 1/3 to raise some good points. But really just read the Nature Explainer.
I think collectively the sources they cite do support their position, or more specifically, they weaken some of the arguments for LabLeak by showing we might expect that evidence even under Zoonosis. The argument summaries:
Pro-Z: (a) Zoonosis has a solid portfolio; (b) There’s way more bats out there than first supposed; (c) There’s way more viruses in them bats; (d) Implicit, but there’s way more bush meat too;
Anti-LL: Key args for LabLeak are almost as likely under zoonosis. Although not mentioned here, that would, alas, include China’s squirreliness.
Based on these I revised my LL forecast from 67% ➛ 61%. The authors put it somewhere below 50%, probably below 10%. Fauci said “very, very, very, very remote possibility”. That seems at most 1:1000, as “remote” is normally <5%. Foretell and Metaculus are about 33%, so I may be too high, but I think we discounted LL too much early on.
Lates NAS newsletter quotes their outgoing editor, Daniel Saarewitz, from 25 years ago:

“The resulting disaffection can fuel social movements that are antagonistic to science and technology.”
Link rot is worse than I thought. blog.ayjay.org/the-rotti…
The full Atlantic article considers some remedies.
Whimsical final slide for great USPTO talk by Wm. Carlson on Tesla & the social process of invention. Notes that 19th C. “Patent Promote Sell” model is the modern startup plan. Also Elon Musk, where “sell” == “license” (and promote is obvious 🙂)

Latest Analytic Insider asks how analysts can help reframe national conversations … around positive narratives:
Suggests the “national security enterprise” write for the general public and be more like:
…Finland where much energy is going into developing positive narratives that take the oxygen away from false narratives
Because
counter[ing] false narratives with facts does not appear to be working. Few supporters of a false narratives will admit they are wrong when presented with “the facts.” …
Toby Handfield mused on a philosopher requesting advice about an upcoming talk:
I don’t know any of the existing literature for this talk, said the visitor, without a hint of embarrassment.
My colleagues earnestly continued to offer advice, not batting an eyelid at this remarkable statement.
This is the wrong response. The visitor had just admitted that he is not competent to give an academic presentation on this topic. He should decline the invitation to speak.
Handfield suggests this reveals a tendency to make philosophy:
something like an elaborate parlour game – in the seminar room we are primarily witnessing a demonstration of cleverness and ingenuity, rather than participating in an ongoing collective enterprise of accumulating knowledge
~ * ~
A thought:One of my Ph.D. advisors frequently incorporated technical material outside of philosophy, as one does in Philosophy of Science. He said that professional philosophers were a difficult audience because they assumed they should be able to absorb and understand any necessary outside material in the course of the seminar. Thought: such folks would likely also feel qualified to present with little preparation. Ego is part of this, but maybe hereditary norms are another.
Famously, some people can do this, at least sometimes. Von Neuman & Feynman spring to mind – the “Feynman Lectures on Computation” for example.[1] I shouldn’t be surprised to hear it of, say, David Lewis. I’ve known a handful who seem close.
If Popper was right that much of philosophy is copying what famous philosophers do, I wonder if the parlour game comes from acting like the top (past) players in the field, when most of us are not. (Or pushing our luck, if we are. )
[1] With Feynman we always have to wonder how much preparation is hidden as part of the trick, but it remains he was highly versatile and quick to absorb new material.
CSET has noted that Chinese and US fighter pilots are losing to AI.
Some of us were doing that back in the 80s.
Arnold Kling: 2021 book titles show epistemological crisis.
This may fit with a historical pattern. The barbarians sack the city, and the carriers of the dying culture repair to their basements to write.
Though some of this goes back at least to Paul Meehl.
HTT: Bob Horn. Again.
Jacobs' Heather Wishart interviews Gen. McChrystal on teams and innovation:
I think our mindset should often be that while we don’t know what we’re going to face, we’re going to develop a team that is really good and an industrial base that is fast. We are going to figure it out as it goes, and we’re going to build to need then. Now, that’s terrifying.
On military acquisition:
The mine-resistant vehicles were a classic case; they didn’t exist at all in the US inventory; we produced thousands of them. [after the war started]
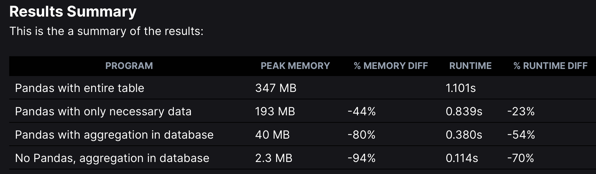
Pandas is wonderful, but Haki Benita reminds us it’s often better to aggregate in the database first:
Dan Miessler reflects on the value of shared culture:
I think young men lacking a shared culture with those around them constitute a national security threat. They are empty vessels waiting to be filled with extremist ideologies around race, religion, and immigration.
(Scroll to end, look for “Declining Religion in the West…")
David Peterson suggests metascience drop its implicit unity-of-science approach:
If reformers want to make diverse scientific fields more robust, they need to demonstrate that they understand how specific fields operate before they push a set of practices about how science overall should operate.